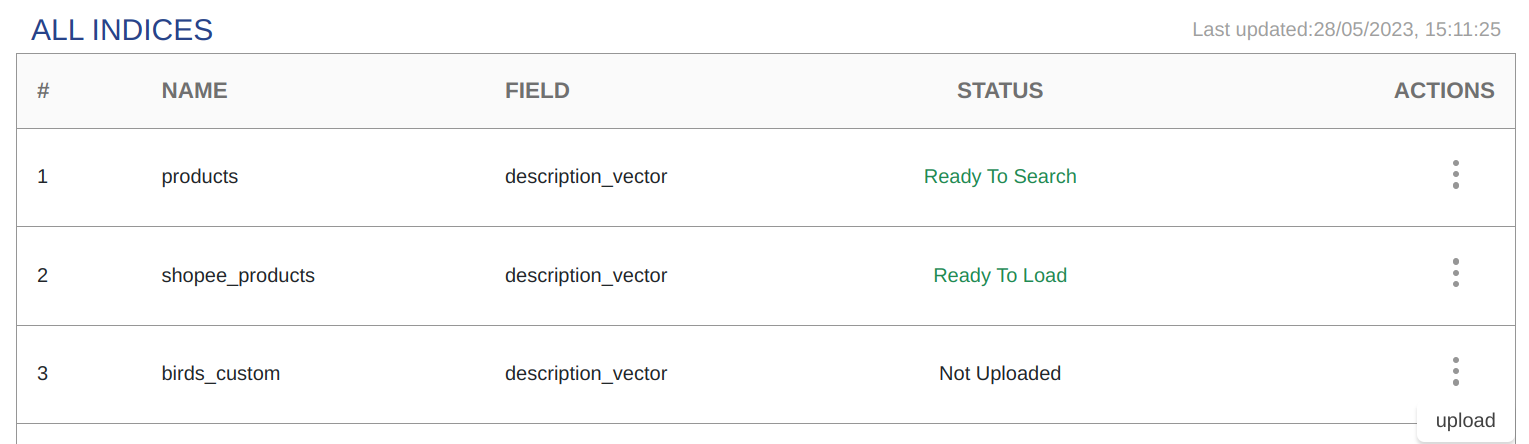
Uploading and Training Indices
Starting the Upload Process
Click the Action icon of an index with a Not Uploaded status and choose Upload from the context menu.
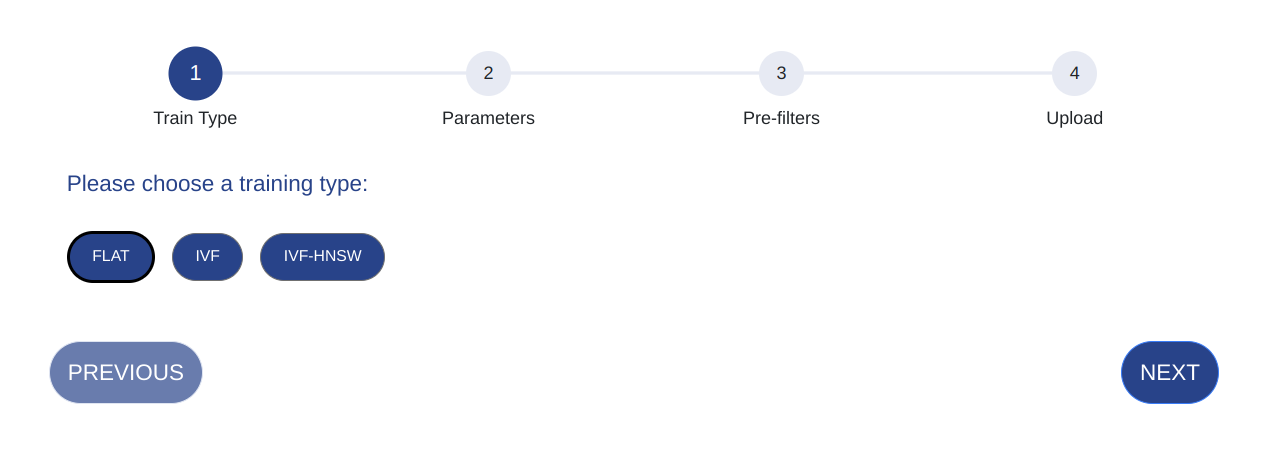
Select the train type for your index. Click NEXT and follow your train type specific uploading instructions.
Flat Uploading
Enter the parameters below. Parameters vary based on training type.
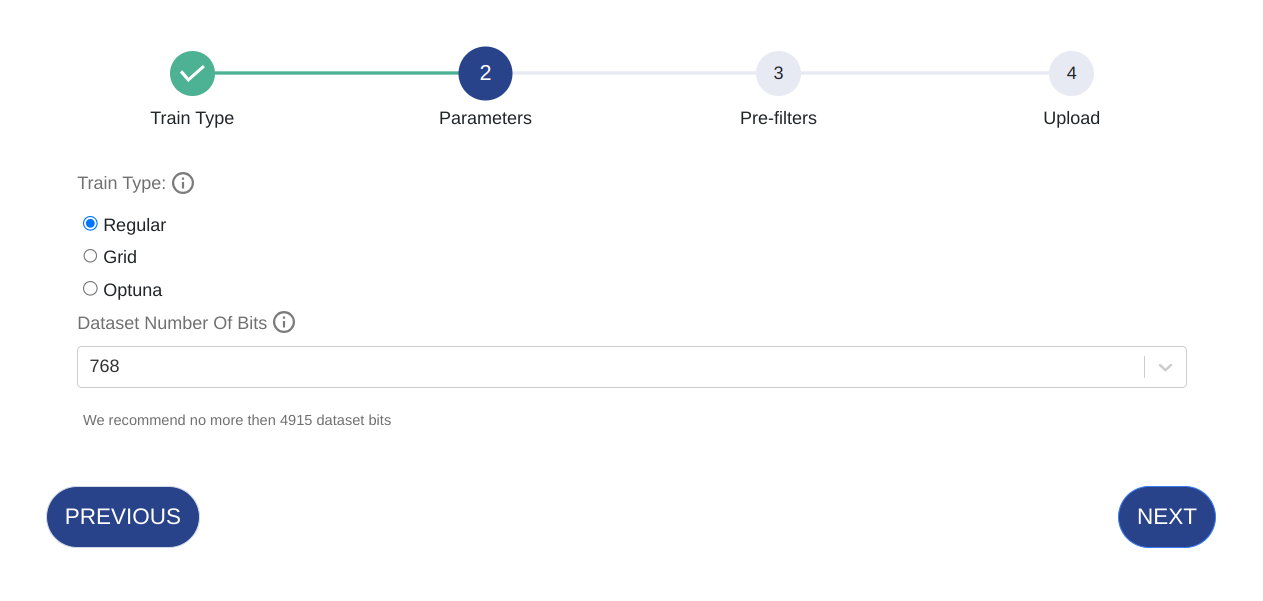
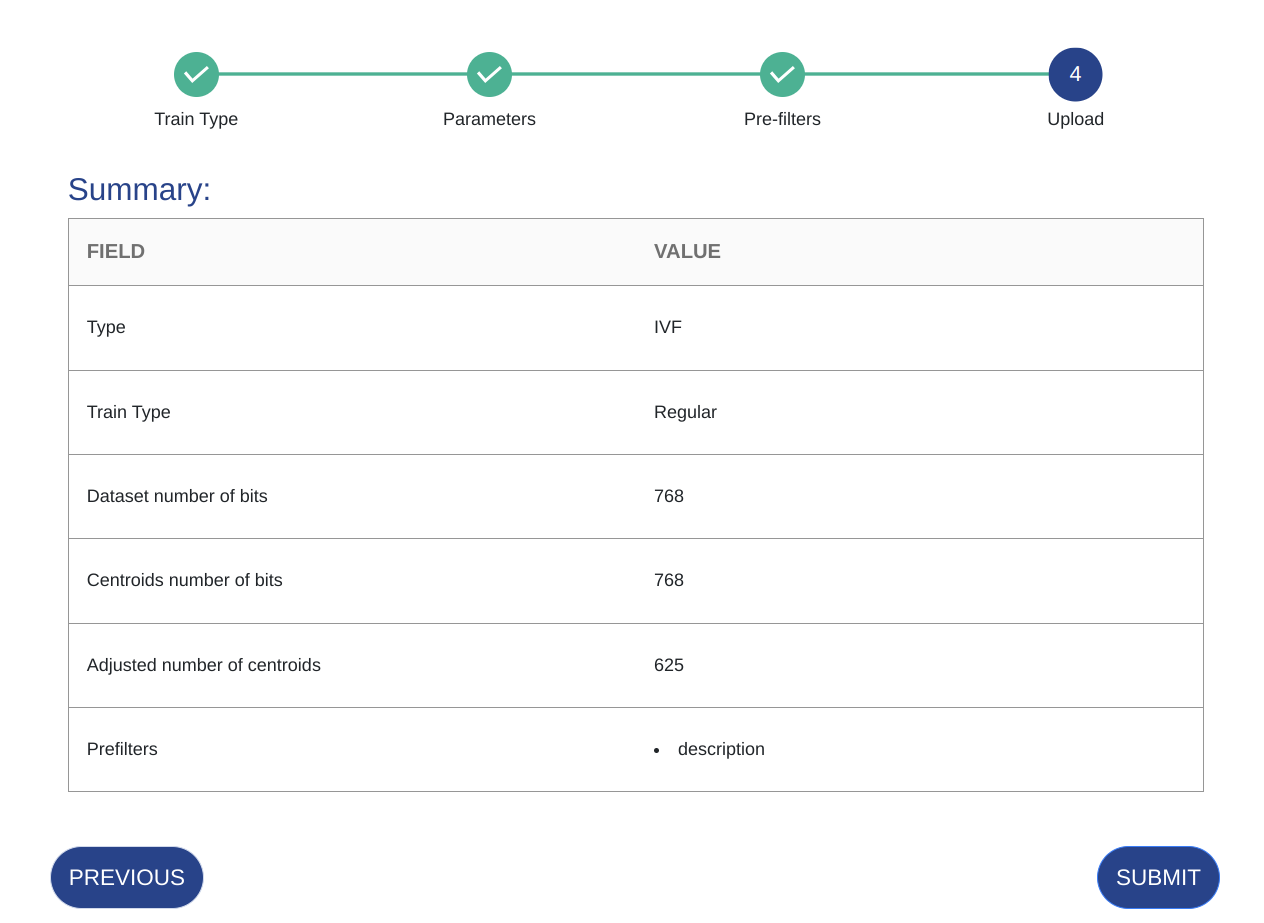
Parameter Details Train Type Optimize the dataset during the train process using Optuna or Grid. Note that this will take longer to train. Dataset N Bits The number of features for which the vector will be in its binary format after quantization.
In the Prefilter window, choose which filters you would like to use when searching through your dataset. Only fields of type
keywordare available as filters.
Verify your choices and click SUBMIT.

IVF Uploading
Enter the parameters below. Parameters vary based on training type.
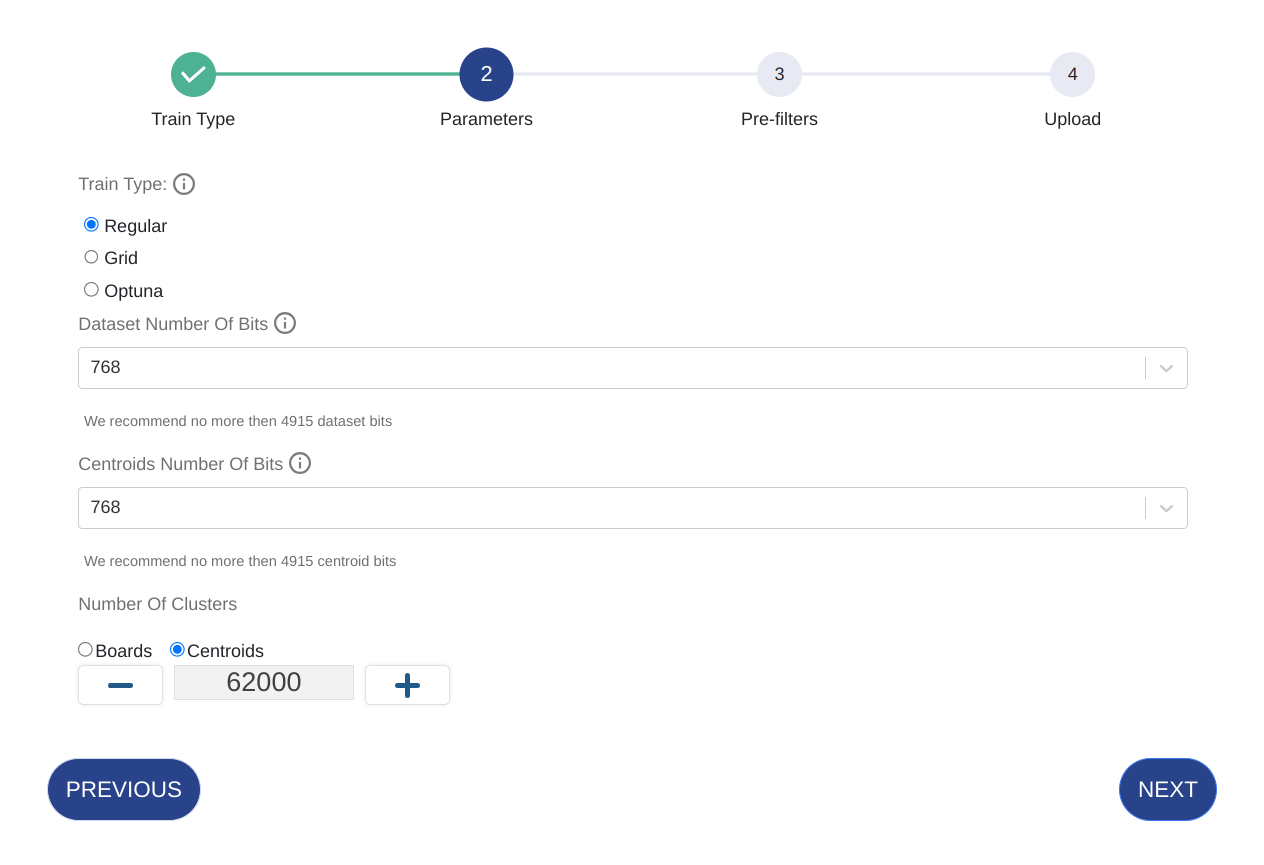
Parameter Details Train Type Optimize the dataset during the train process using Optuna or Grid. Note that this will take longer to train. Dataset N Bits The number of features for which the vector will be in its binary format after quantization. Cluster N Bits The number of features for which the centroids vectors will be in their binary format after quantization Number of Clusters The number of clusters in the IFV structure. If you select Boards, then the number of clusters will be automatically determined based on the maximum possible.
In the Prefilter window, choose which filters you would like to use when searching through your dataset. Only fields of type
keywordare available as filters.
Verify your choices and click SUBMIT.
IVF-HNSW Uploading
Enter the parameters below. Parameters vary based on training type.
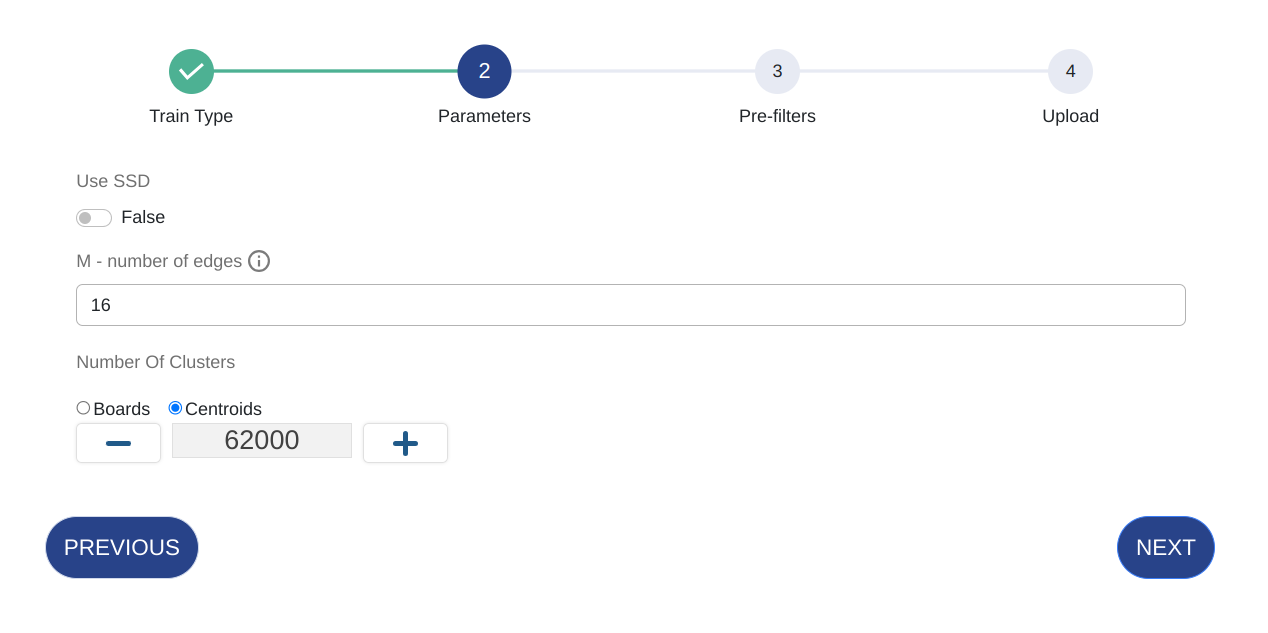
Parameter Details Use SSD Use SSD for memory efficiency. M - number of edges Equivalent to m. The number of neighbors for each node in the HNSW graph.Number of Clusters Equivalent to ef_construction. The number of clusters in the IFV-HNSW structure. If you select Boards, then the number of clusters will be automatically determined based on the maximum possible.
In the Prefilter window, choose which filters you would like to use when searching through your dataset. Only fields of type
keywordare available as filters.
Verify your choices and click SUBMIT.
